3.3 Allotriploid C. insueta
3.3.1 Introduction
Allotriploid Cardamine insueta (2n = 3x = 24, RRA) serves as an example of an uneven subgenome composition. Two of its subgenomes are derived from Cardamine rivularis (2n = 2x = 16, RR), a species inhabiting grassy areas away from riversides, while the third subgenome comes from Cardamine amara (2n = 2x = 16, AA), a species typically found on riversides or in streams (Urbanska-Worytkiewicz and Landolt 1972, 1974). RNA-Seq studies on C. insueta leaflets floating on water suggested that C. insueta shifts homeolog expression ratios over time following submergence (Sun et al. 2020).
3.3.2 Data Preparation
Here, we demonstrate how to detect homeologs with changing expression ratios across time points using HOBIT,
similar to an ANOVA approach.
We use a sample dataset containing 500 homeolog pairs (c_insueta.mini.txt.gz) randomly selected from the original experiment,
although the full dataset is available in the data directory
for users who wish to analyze it.
This data span nine time points (0, 2, 4, 8, 12, 24, 48, 72, and 96 hours)
following leaf submergence and can be loaded using the following code.
gexp <- read.table("../data/c_insueta.mini.txt.gz",
header = TRUE, sep = "\t", row.names = 1)
head(gexp)## T00 T02 T04 T08 T12 T24 T48 T72 T96
## CARHR000430_R 148 32 134 76 118 54 119 144 94
## CARHR000770_R 88 127 17 2 56 25 35 22 17
## CARHR001290_R 52 18 84 35 63 12 33 43 34
## CARHR001700_R 48 15 29 29 71 35 62 47 35
## CARHR001770_R 12 5 18 14 22 8 150 7 8
## CARHR001840_R 108 20 129 58 118 52 109 163 82Next, load the homeolog mapping table, which links gene expression values to their corresponding homeolog pairs. This file is tab-separated, with the first and second columns representing genes from C. rivularis and C. amara, respectively.
mapping_table <- read.table("../data/c_insueta.homeolog.txt.gz",
header = TRUE, sep = "\t")
head(mapping_table)## C_rivularis C_amara
## 1 CARHR000010_R CARHR000010_A
## 2 CARHR000060_R CARHR000060_A
## 3 CARHR000090_R CARHR000090_A
## 4 CARHR000110_R CARHR000110_A
## 5 CARHR000120_R CARHR000120_A
## 6 CARHR000130_R CARHR000130_AUse the newExpMX() function
to create a homeolog expression matrix from the gene expression data (gexp)
and mapping table (mapping_table).
The result is stored as an ExpMX class object.
## # 2 subgenome sets (C_rivularis, C_amara)
## # 500 homeolog tuples
## ---------------------
## Experiment Design:
## group
## 1 T00
## 2 T02
## 3 T04
## 4 T08
## 5 T12
## 6 T24
## 7 T48
## 8 T72
## 9 T96
## ---------------------
## > subgenome: C_rivularis
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9]
## [1,] 148 32 134 76 118 54 119 144 94
## [2,] 88 127 17 2 56 25 35 22 17
## [3,] 52 18 84 35 63 12 33 43 34
## [4,] 48 15 29 29 71 35 62 47 35
## [5,] 12 5 18 14 22 8 150 7 8
## [6,] 108 20 129 58 118 52 109 163 82
## +++++++++++++++++++++
## > subgenome: C_amara
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9]
## [1,] 45 15 42 34 61 34 47 44 38
## [2,] 30 64 10 21 54 17 24 15 11
## [3,] 20 13 54 25 26 9 23 18 12
## [4,] 23 10 19 7 21 8 31 20 12
## [5,] 6 2 2 5 16 5 85 1 3
## [6,] 48 10 74 16 68 36 56 67 52
## ---------------------Normalize the read counts using the TMM method (Robinson and Oshlack 2010), since the data has not been normalized.
Users can visualize the distribution of homeolog expression ratios using the plot_HER_distr() function.
By default, the expression ratio is calculated as
the proportion contributed by the first subgenome relative to all subgenomes,
where the first subgenome corresponds to C. rivularis,
as listed in the first column of the mapping table (mapping_table).
The plot_HER_distr() function generates a histogram for each time point.
distr_plots <- plot_HER_distr(x)
library(ggplot2)
library(gridExtra)
distr_plots <- lapply(seq_along(distr_plots), function(p) {
distr_plots[[p]] + ggtitle(names(distr_plots)[p])
})
grid.arrange(grobs = distr_plots, ncol = 3)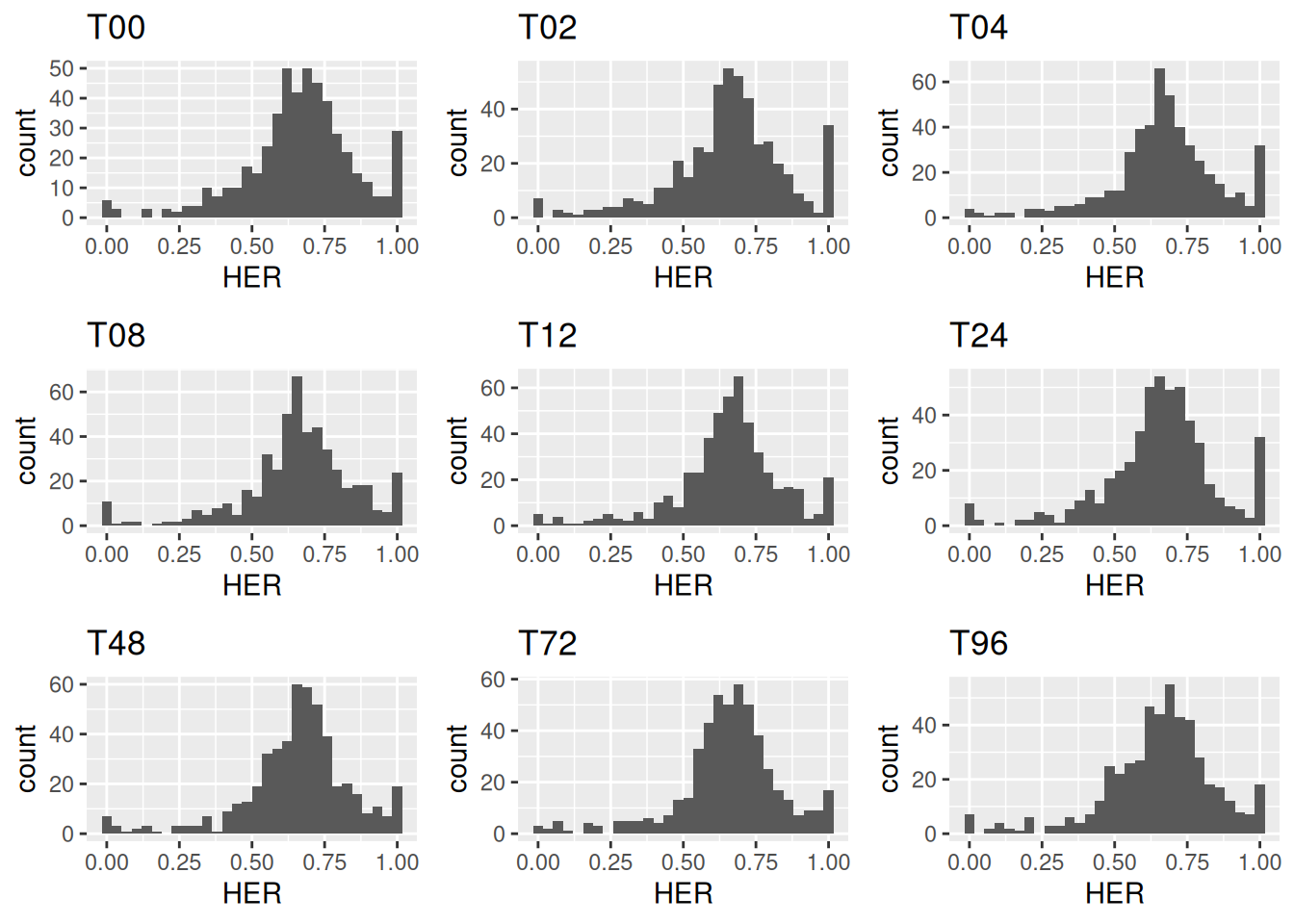
Figure 3.9: Distribution of homeolog expression ratios in C. insueta across nine time points following leaf submergence in water.
These visualizations indicate that most homeologs in C. insueta are expressed at a 2:1 ratio between the C. rivularis and C. amara subgenomes, as shown by histogram peaks centered around two-thirds.
3.3.3 Statistical Test
Next, apply the hobit() function
to detect homeologs with differential expression ratios across the nine time points following leaf submergence.
Since this dataset contains only one replicate per condition,
set no_replicate = TRUE to avoid errors arising from dispersion estimation,
which requires multiple replicates.
This step takes approximately five minutes using eight threads and may vary depending on hardware performance.
## gene pvalue qvalue raw_pvalue raw_qvalue D__C_rivularis__(T00-T02) D__C_rivularis__(T00-T04) D__C_rivularis__(T00-T08) D__C_rivularis__(T00-T12) D__C_rivularis__(T00-T24) D__C_rivularis__(T00-T48) D__C_rivularis__(T00-T72) D__C_rivularis__(T00-T96) D__C_rivularis__(T02-T04) D__C_rivularis__(T02-T08) D__C_rivularis__(T02-T12) D__C_rivularis__(T02-T24) D__C_rivularis__(T02-T48) D__C_rivularis__(T02-T72) D__C_rivularis__(T02-T96) D__C_rivularis__(T04-T08) D__C_rivularis__(T04-T12) D__C_rivularis__(T04-T24) D__C_rivularis__(T04-T48) D__C_rivularis__(T04-T72) D__C_rivularis__(T04-T96) D__C_rivularis__(T08-T12) D__C_rivularis__(T08-T24) D__C_rivularis__(T08-T48) D__C_rivularis__(T08-T72) D__C_rivularis__(T08-T96) D__C_rivularis__(T12-T24) D__C_rivularis__(T12-T48) D__C_rivularis__(T12-T72) D__C_rivularis__(T12-T96) D__C_rivularis__(T24-T48) D__C_rivularis__(T24-T72) D__C_rivularis__(T24-T96) D__C_rivularis__(T48-T72) D__C_rivularis__(T48-T96) D__C_rivularis__(T72-T96) D__C_amara__(T00-T02) D__C_amara__(T00-T04) D__C_amara__(T00-T08) D__C_amara__(T00-T12) D__C_amara__(T00-T24) D__C_amara__(T00-T48) D__C_amara__(T00-T72) D__C_amara__(T00-T96) D__C_amara__(T02-T04) D__C_amara__(T02-T08) D__C_amara__(T02-T12) D__C_amara__(T02-T24) D__C_amara__(T02-T48) D__C_amara__(T02-T72) D__C_amara__(T02-T96) D__C_amara__(T04-T08) D__C_amara__(T04-T12) D__C_amara__(T04-T24) D__C_amara__(T04-T48) D__C_amara__(T04-T72) D__C_amara__(T04-T96) D__C_amara__(T08-T12) D__C_amara__(T08-T24) D__C_amara__(T08-T48) D__C_amara__(T08-T72) D__C_amara__(T08-T96) D__C_amara__(T12-T24) D__C_amara__(T12-T48) D__C_amara__(T12-T72) D__C_amara__(T12-T96) D__C_amara__(T24-T48) D__C_amara__(T24-T72) D__C_amara__(T24-T96) D__C_amara__(T48-T72) D__C_amara__(T48-T96) D__C_amara__(T72-T96) OR__C_rivularis__(T00/T02) OR__C_rivularis__(T00/T04) OR__C_rivularis__(T00/T08) OR__C_rivularis__(T00/T12) OR__C_rivularis__(T00/T24) OR__C_rivularis__(T00/T48) OR__C_rivularis__(T00/T72) OR__C_rivularis__(T00/T96) OR__C_rivularis__(T02/T04) OR__C_rivularis__(T02/T08) OR__C_rivularis__(T02/T12) OR__C_rivularis__(T02/T24) OR__C_rivularis__(T02/T48) OR__C_rivularis__(T02/T72) OR__C_rivularis__(T02/T96) OR__C_rivularis__(T04/T08) OR__C_rivularis__(T04/T12) OR__C_rivularis__(T04/T24) OR__C_rivularis__(T04/T48) OR__C_rivularis__(T04/T72) OR__C_rivularis__(T04/T96) OR__C_rivularis__(T08/T12) OR__C_rivularis__(T08/T24) OR__C_rivularis__(T08/T48) OR__C_rivularis__(T08/T72) OR__C_rivularis__(T08/T96) OR__C_rivularis__(T12/T24) OR__C_rivularis__(T12/T48) OR__C_rivularis__(T12/T72) OR__C_rivularis__(T12/T96) OR__C_rivularis__(T24/T48) OR__C_rivularis__(T24/T72) OR__C_rivularis__(T24/T96) OR__C_rivularis__(T48/T72) OR__C_rivularis__(T48/T96) OR__C_rivularis__(T72/T96) OR__C_amara__(T00/T02) OR__C_amara__(T00/T04) OR__C_amara__(T00/T08) OR__C_amara__(T00/T12) OR__C_amara__(T00/T24) OR__C_amara__(T00/T48) OR__C_amara__(T00/T72) OR__C_amara__(T00/T96) OR__C_amara__(T02/T04) OR__C_amara__(T02/T08) OR__C_amara__(T02/T12) OR__C_amara__(T02/T24) OR__C_amara__(T02/T48) OR__C_amara__(T02/T72) OR__C_amara__(T02/T96) OR__C_amara__(T04/T08) OR__C_amara__(T04/T12) OR__C_amara__(T04/T24) OR__C_amara__(T04/T48) OR__C_amara__(T04/T72) OR__C_amara__(T04/T96) OR__C_amara__(T08/T12) OR__C_amara__(T08/T24) OR__C_amara__(T08/T48) OR__C_amara__(T08/T72) OR__C_amara__(T08/T96) OR__C_amara__(T12/T24) OR__C_amara__(T12/T48) OR__C_amara__(T12/T72) OR__C_amara__(T12/T96) OR__C_amara__(T24/T48) OR__C_amara__(T24/T72) OR__C_amara__(T24/T96) OR__C_amara__(T48/T72) OR__C_amara__(T48/T96) OR__C_amara__(T72/T96) Dmax ORmax theta0__C_rivularis theta0__C_amara theta1__C_rivularis__T00 theta1__C_amara__T00 theta1__C_rivularis__T02 theta1__C_amara__T02 theta1__C_rivularis__T04 theta1__C_amara__T04 theta1__C_rivularis__T08 theta1__C_amara__T08 theta1__C_rivularis__T12 theta1__C_amara__T12 theta1__C_rivularis__T24 theta1__C_amara__T24 theta1__C_rivularis__T48 theta1__C_amara__T48 theta1__C_rivularis__T72 theta1__C_amara__T72 theta1__C_rivularis__T96 theta1__C_amara__T96 logLik_H0 logLik_H1
## 1 CARHR000430_R 1.0000000 1 1.0000000 1 0.07390725 0.004128505 0.07188315 0.097352840 0.13871523 0.042145770 -0.002880205 0.044821730 -0.068730000 -0.010411110 0.02223977 0.063146665 -0.033699705 -0.08126181 -0.030565685 0.06560613 0.09599315 0.14023605 0.04209613 -0.007845965 0.04312116 0.02716275 0.068061075 -0.030448465 -0.07588518 -0.02286770 0.04033711 -0.06029592 -0.099953195 -0.04932497 -0.09689848 -0.143894025 -0.092220115 -0.04629152 0.00714313 0.05687011 -0.07390725 -0.004128505 -0.07188315 -0.097352840 -0.13871523 -0.042145770 0.002880205 -0.044821730 0.068730000 0.010411110 -0.02223977 -0.063146665 0.033699705 0.08126181 0.030565685 -0.06560613 -0.09599315 -0.14023605 -0.04209613 0.007845965 -0.04312116 -0.02716276 -0.068061075 0.030448465 0.07588518 0.02286771 -0.04033712 0.06029593 0.099953195 0.04932498 0.09689848 0.143894025 0.092220115 0.046291520 -0.00714313 -0.05687011 1.4495637 1.0258950 1.4511202 1.627020 1.9617633 1.2404358 0.9836465 1.2599861 0.7078819 0.9514909 1.1100515 1.3307903 0.8520311 0.6638057 0.8658834 1.3873184 1.5994232 1.9371656 1.2413136 0.9582132 1.2512546 1.1390013 1.3621298 0.8631400 0.6778681 0.8944764 1.1941450 0.7507847 0.6069116 0.7949455 0.6326680 0.5040128 0.6608710 0.7809329 1.0341152 1.3487126 0.6898627 0.9747624 0.6891229 0.6146208 0.5097455 0.8061695 1.0166255 0.7936596 1.4126652 1.0509822 0.9008592 0.7514337 1.1736661 1.5064650 1.1548899 0.7208151 0.6252254 0.5162181 0.8055982 1.0436093 0.7991979 0.8779621 0.7341444 1.1585606 1.4752132 1.1179725 0.8374193 1.3319398 1.6476864 1.2579479 1.5806078 1.9840767 1.5131546 1.2805199 0.9670103 0.7414480 0.1438940 1.984077 0.6712120 0.3287880 0.7400893 0.2599107 0.6603915 0.3396085 0.7351254 0.2648746 0.6646895 0.3353105 0.6357553 0.3642447 0.5925513 0.4074487 0.6931491 0.3068509 0.7457773 0.2542227 0.6902934 0.3097066 -82.66770 -84.60827
## 2 CARHR000770_R 0.7889875 1 0.7889875 1 0.04884087 0.028309735 0.35479750 0.117691785 0.06509919 0.060027045 0.063671440 0.057782765 -0.018988890 0.291309250 0.05478218 0.009201175 0.008252530 0.01171802 0.007231095 0.30665859 0.07667807 0.02644064 0.02771935 0.026054045 0.02979770 -0.22445893 -0.262611390 -0.274166634 -0.27600382 -0.27019951 -0.05047256 -0.04841941 -0.044715915 -0.05212607 0.00162436 0.001615705 -0.006995785 0.00274261 -0.00105112 0.00702302 -0.04884087 -0.028309735 -0.35479750 -0.117691787 -0.06509919 -0.060027045 -0.063671440 -0.057782765 0.018988890 -0.291309250 -0.05478218 -0.009201175 -0.008252531 -0.01171802 -0.007231095 -0.30665860 -0.07667807 -0.02644063 -0.02771935 -0.026054042 -0.02979769 0.22445894 0.262611390 0.274166635 0.27600382 0.27019951 0.05047257 0.04841941 0.044715913 0.05212607 -0.00162436 -0.001615705 0.006995784 -0.002742611 0.00105112 -0.00702302 1.3582991 1.2080634 7.0433868 1.940977 1.4875404 1.4610572 1.4771289 1.4249944 0.9060193 4.7074449 1.3282066 1.0537122 1.0545797 1.0730280 1.0442989 5.6387122 1.5379193 1.1639210 1.1857070 1.1618275 1.1923401 0.2787721 0.2286357 0.2128642 0.2097411 0.2144983 0.7584886 0.7685926 0.7752671 0.7534706 1.0107634 1.0077592 0.9565796 1.0158057 0.9950660 1.0463373 0.7362151 0.8277711 0.1419772 0.5152061 0.6722548 0.6844365 0.6769894 0.7017691 1.1037304 0.2124299 0.7528949 0.9490257 0.9482453 0.9319425 0.9575803 0.1773455 0.6502293 0.8591653 0.8433788 0.8607132 0.8386869 3.5871600 4.3737705 4.6978412 4.7678021 4.6620416 1.3184114 1.3010803 1.2898791 1.3271918 0.9893524 0.9923006 1.0454012 0.9844407 1.0049587 0.9557154 0.3547975 7.043387 0.5713192 0.4286808 0.7103487 0.2896513 0.6380701 0.3619299 0.6569026 0.3430974 0.2651404 0.7348596 0.5594410 0.4405590 0.6242456 0.3757544 0.6284016 0.3715984 0.6200014 0.3799986 0.6258876 0.3741124 -92.60451 -90.07036
## 3 CARHR001290_R 1.0000000 1 1.0000000 1 0.11097877 0.098611155 0.11286804 0.001558095 0.11442781 0.102048140 0.011505765 -0.008719305 -0.008932075 0.001582130 -0.10213876 0.004865270 -0.005310995 -0.09528440 -0.126167450 0.01507699 -0.09163656 0.01081457 0.00586452 -0.085800405 -0.11201532 -0.10298059 0.003248270 -0.007716745 -0.09715092 -0.12536081 0.10382148 0.09772689 0.006788715 -0.02722386 -0.01795330 -0.098128385 -0.129225465 -0.08853248 -0.12127331 -0.02536999 -0.11097878 -0.098611157 -0.11286804 -0.001558095 -0.11442780 -0.102048140 -0.011505765 0.008719305 0.008932075 -0.001582130 0.10213876 -0.004865270 0.005310995 0.09528440 0.126167450 -0.01507699 0.09163656 -0.01081458 -0.00586452 0.085800405 0.11201532 0.10298059 -0.003248270 0.007716745 0.09715092 0.12536082 -0.10382148 -0.09772688 -0.006788715 0.02722386 0.01795330 0.098128385 0.129225465 0.088532485 0.12127331 0.02537000 1.6938850 1.5806422 1.7114933 1.009104 1.6972393 1.6304543 1.0599082 0.9580539 0.9611638 1.0067618 0.6184725 1.0207787 0.9766388 0.6481428 0.5505337 1.0703951 0.6562603 1.0551142 1.0336242 0.6694582 0.5779524 0.6143103 1.0136825 0.9664135 0.6367689 0.5440377 1.6044944 1.5916576 1.0361645 0.8614407 0.9252113 0.6359783 0.5447083 0.6622428 0.5549399 0.8744945 0.5903588 0.6326542 0.5842852 0.9909785 0.5891924 0.6133266 0.9434781 1.0437826 1.0404055 0.9932838 1.6168868 0.9796443 1.0239202 1.5428698 1.8164200 0.9342356 1.5237855 0.9477649 0.9674703 1.4937454 1.7302465 1.6278434 0.9865022 1.0347539 1.5704285 1.8381079 0.6232493 0.6282759 0.9650977 1.1608460 1.0808341 1.5723808 1.8358453 1.5100203 1.8019969 1.1435196 0.1292255 1.838108 0.5967689 0.4032311 0.6768891 0.3231109 0.5552771 0.4447229 0.5706934 0.4293066 0.5590459 0.4409541 0.6715994 0.3284006 0.5561963 0.4438037 0.5655119 0.4344881 0.6642194 0.3357806 0.6950216 0.3049784 -76.36424 -77.69972
## 4 CARHR001700_R 1.0000000 1 1.0000000 1 0.06647638 0.058106730 -0.12453312 -0.090393675 -0.13278630 0.010317885 -0.021532695 -0.055224010 -0.006773340 -0.184664405 -0.15165196 -0.196262465 -0.056055760 -0.08514081 -0.120882550 -0.18200308 -0.14747757 -0.18989757 -0.05005963 -0.078285270 -0.11621813 0.03398578 -0.006656665 0.130320775 0.10643994 0.06547072 -0.04326171 0.09589544 0.072122640 0.03073139 0.14021238 0.113916090 0.076165075 -0.02521588 -0.06410192 -0.03920024 -0.06647638 -0.058106730 0.12453311 0.090393675 0.13278630 -0.010317885 0.021532695 0.055224010 0.006773340 0.184664405 0.15165196 0.196262465 0.056055760 0.08514081 0.120882550 0.18200309 0.14747756 0.18989757 0.05005964 0.078285270 0.11621813 -0.03398578 0.006656665 -0.130320775 -0.10643995 -0.06547072 0.04326170 -0.09589544 -0.072122640 -0.03073139 -0.14021238 -0.113916090 -0.076165075 0.025215885 0.06410192 0.03920024 1.3427424 1.3097379 0.5039799 0.634801 0.4830749 1.0506825 0.9024424 0.7608741 0.9709373 0.3905760 0.4760649 0.3611154 0.7731732 0.6807675 0.5617244 0.3941841 0.4842173 0.3741673 0.8017260 0.6940608 0.5802253 1.2260328 0.9586462 2.0243934 1.7938502 1.4598359 0.7551644 1.6376205 1.4596738 1.1809248 2.1495686 1.8978022 1.5682674 0.8823165 0.7257262 0.8159309 0.7447445 0.7635116 1.9842062 1.5752970 2.0700724 0.9517623 1.1081041 1.3142779 1.0299327 2.5603214 2.1005540 2.7691980 1.2933717 1.4689305 1.7802326 2.5368861 2.0651887 2.6726017 1.2473089 1.4407961 1.7234686 0.8156389 1.0431385 0.4939751 0.5574602 0.6850085 1.3242155 0.6106421 0.6850846 0.8467940 0.4652096 0.5269254 0.6376463 1.1333812 1.3779307 1.2255944 0.1962625 2.769198 0.6861603 0.3138397 0.6654042 0.3345958 0.5983534 0.4016466 0.6024057 0.3975943 0.7927729 0.2072271 0.7582178 0.2417822 0.8036509 0.1963491 0.6557708 0.3442292 0.6835962 0.3164038 0.7253547 0.2746453 -69.05419 -69.67870
## 5 CARHR001770_R 0.8862251 1 0.8862251 1 -0.02791533 -0.150907360 -0.04340837 0.038030910 0.01947772 -0.005324557 -0.168933605 -0.047135527 -0.112251500 -0.005145935 0.06453639 0.061678386 0.023656925 -0.12690875 -0.020180775 0.10922586 0.19588738 0.17733095 0.15489058 -0.015997525 0.10304881 0.07870959 0.046109840 0.046788658 -0.13005074 -0.00918908 -0.01704995 -0.03670061 -0.213736905 -0.07917805 -0.02476112 -0.200230105 -0.071691270 -0.17032771 -0.05014754 0.12832122 0.02791534 0.150907360 0.04340837 -0.038030910 -0.01947772 0.005324555 0.168933602 0.047135525 0.112251500 0.005145935 -0.06453639 -0.061678385 -0.023656925 0.12690875 0.020180775 -0.10922586 -0.19588738 -0.17733095 -0.15489058 0.015997525 -0.10304881 -0.07870959 -0.046109840 -0.046788660 0.13005074 0.00918908 0.01704994 0.03670061 0.213736905 0.07917805 0.02476112 0.200230105 0.071691270 0.170327703 0.05014754 -0.12832122 0.8558299 0.3979026 0.7857015 1.237595 1.1035543 0.9755351 0.3357484 0.7777813 0.4866563 0.9777441 1.4183158 1.3874280 1.1481049 0.4063571 0.8917662 2.0522899 3.1049158 2.8613756 2.5012422 0.8525692 1.9660261 1.5439420 1.2996599 1.2790493 0.4270281 0.9515674 0.9123741 0.8163818 0.2855157 0.6520266 0.8711677 0.3011388 0.6604579 0.3593655 0.7594885 2.3906859 1.1684585 2.5131783 1.2727511 0.8080191 0.9061629 1.0250786 2.9784219 1.2857129 2.0548393 1.0227628 0.7050632 0.7207584 0.8710006 2.4608899 1.1213715 0.4872616 0.3220702 0.3494836 0.3998023 1.1729264 0.5086403 0.6476929 0.7694329 0.7818311 2.3417682 1.0508981 1.0960417 1.2249171 3.5024379 1.5336898 1.1478847 3.3207332 1.5141011 2.7826841 1.3166756 0.4182900 0.2137369 3.502438 0.5640001 0.4359999 0.5317814 0.4682186 0.5863351 0.4136649 0.7406336 0.2593664 0.5872280 0.4127720 0.4849471 0.5150529 0.5091254 0.4908746 0.5364667 0.4635333 0.7658730 0.2341270 0.5964340 0.4035660 -73.03252 -70.93236
## 6 CARHR001840_R 1.0000000 1 1.0000000 1 0.01853578 0.056611105 -0.08534611 0.051690805 0.08954754 0.031126685 -0.019049805 0.073248140 0.031716240 -0.106348225 0.03441213 0.076494890 0.007634035 -0.03577025 0.055223220 -0.14048567 -0.00618485 0.03799053 -0.02828861 -0.071302515 0.01891534 0.14037748 0.176768775 0.116194705 0.07129573 0.16035196 0.04119422 -0.02138005 -0.060656165 0.02078637 -0.05955605 -0.104947945 -0.014759730 -0.04430433 0.04501858 0.08937892 -0.01853578 -0.056611105 0.08534611 -0.051690805 -0.08954753 -0.031126685 0.019049805 -0.073248140 -0.031716240 0.106348225 -0.03441213 -0.076494890 -0.007634035 0.03577025 -0.055223220 0.14048567 0.00618485 -0.03799054 0.02828861 0.071302515 -0.01891534 -0.14037748 -0.176768775 -0.116194705 -0.07129573 -0.16035195 -0.04119421 0.02138005 0.060656165 -0.02078637 0.05955605 0.104947945 0.014759730 0.044304330 -0.04501857 -0.08937892 1.0930823 1.3039117 0.6381697 1.256836 1.4890041 1.1569077 0.9144944 1.3888264 1.1524719 0.5752922 1.1696735 1.4026222 1.0361399 0.8405345 1.2784183 0.4935254 0.9718433 1.1812344 0.8815919 0.7169366 1.0889022 2.0139153 2.3426045 1.8188593 1.4463163 2.1627806 1.1976225 0.9091645 0.7558396 1.0995389 0.7691875 0.6227997 0.9387528 0.8107497 1.2233059 1.4997546 0.9148442 0.7669231 1.5669813 0.7956491 0.6715898 0.8643732 1.0935004 0.7200328 0.8677002 1.7382477 0.8549396 0.7129506 0.9651208 1.1897192 0.7822171 2.0262392 1.0289725 0.8465733 1.1343119 1.3948238 0.9183562 0.4965460 0.4268753 0.5497953 0.6914117 0.4623679 0.8349877 1.0999110 1.3230321 0.9094722 1.3000732 1.6056532 1.0652433 1.2334262 0.8174588 0.6667758 0.1767688 2.342604 0.6300791 0.3699209 0.6687567 0.3312433 0.6475398 0.3524602 0.6088712 0.3911288 0.7585514 0.2414486 0.6121381 0.3878619 0.5712568 0.4287432 0.6365237 0.3634763 0.6830978 0.3169022 0.5893123 0.4106877 -83.81416 -85.597273.3.4 Analysis Environment
## R version 4.5.1 (2025-06-13)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.3 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8 LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8
## [6] LC_MESSAGES=C.UTF-8 LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] UpSetR_1.4.0 gridExtra_2.3 ggplot2_4.0.0 hespresso_1.0.1
##
## loaded via a namespace (and not attached):
## [1] tensorA_0.36.2.1 sass_0.4.10 future_1.67.0 generics_0.1.4 lattice_0.22-7
## [6] hms_1.1.3 listenv_0.9.1 digest_0.6.37 magrittr_2.0.4 evaluate_1.0.5
## [11] grid_4.5.1 RColorBrewer_1.1-3 bookdown_0.44 fastmap_1.2.0 plyr_1.8.9
## [16] jsonlite_2.0.0 processx_3.8.6 progress_1.2.3 backports_1.5.0 limma_3.64.3
## [21] ps_1.9.1 scales_1.4.0 codetools_0.2-20 jquerylib_0.1.4 abind_1.4-8
## [26] cli_3.6.5 rlang_1.1.6 crayon_1.5.3 cmdstanr_0.9.0 parallelly_1.45.1
## [31] future.apply_1.20.0 splines_4.5.1 withr_3.0.2 cachem_1.1.0 yaml_2.3.10
## [36] tools_4.5.1 parallel_4.5.1 checkmate_2.3.3 locfit_1.5-9.12 globals_0.18.0
## [41] vctrs_0.6.5 posterior_1.6.1 R6_2.6.1 lifecycle_1.0.4 edgeR_4.6.3
## [46] pkgconfig_2.0.3 progressr_0.16.0 pillar_1.11.1 bslib_0.9.0 gtable_0.3.6
## [51] Rcpp_1.1.0 data.table_1.17.8 glue_1.8.0 statmod_1.5.0 xfun_0.53
## [56] tibble_3.3.0 knitr_1.50 farver_2.1.2 htmltools_0.5.8.1 labeling_0.4.3
## [61] rmarkdown_2.29 compiler_4.5.1 prettyunits_1.2.0 S7_0.2.0 distributional_0.5.0